Sponsored by NIH – General Medicine
Principle Investigator: Paul G. Falkowski
As large amounts of sequence information become available from the Human Genome Project, sequence analyses will provide clearer evidence that numerous human diseases or disorders have deep evolutionary roots. The project addresses the evolutionary origins of proteases and cell death responses as a case in point. Specifically, the research seeks to:
- examine to the evolutionary origins of autocatalyzed cell death associated with the stress induced expression of novel (cryptic) proteases in single-celled, asexual eucaryotes, specifically the obligate photoautotrophic chlorophyte alga, Dunaliella tertiolecta,
- elucidate the molecular triggers in physiological stress responses that lead to the induction of novel proteases and catastrophic cell death in such cells, and
- examine to what extent the cryptic proteases are ubiquitous in eucaryotes.
These goals address the biological significance of apoptosis and autocatalyzed cell death in the context of the evolution of somatic eucaryotes and the processes that repress or silence the expression of cryptic proteases encoded within their nuclear genomes.
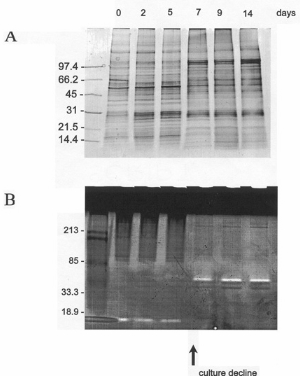
In humans, protease-triggered apoptosis occurs in cytotoxic T lymphocytes and other defensive cell lines, but is also a symptom of numerous chronic diseases or disorders with little or no clear Mendalian genetic lineage. We hypothesize that the genes encoding the stress-induced proteases have been incorporated into bacterial and eukaryotic genomes from relic viral infections, comparable to the endogenous retroviral protease genes found in metazoans. These genes, normally silenced either by repressor factors or transpositions, appear to be derepressed when the organism is selectively stressed.
